Coller Lab

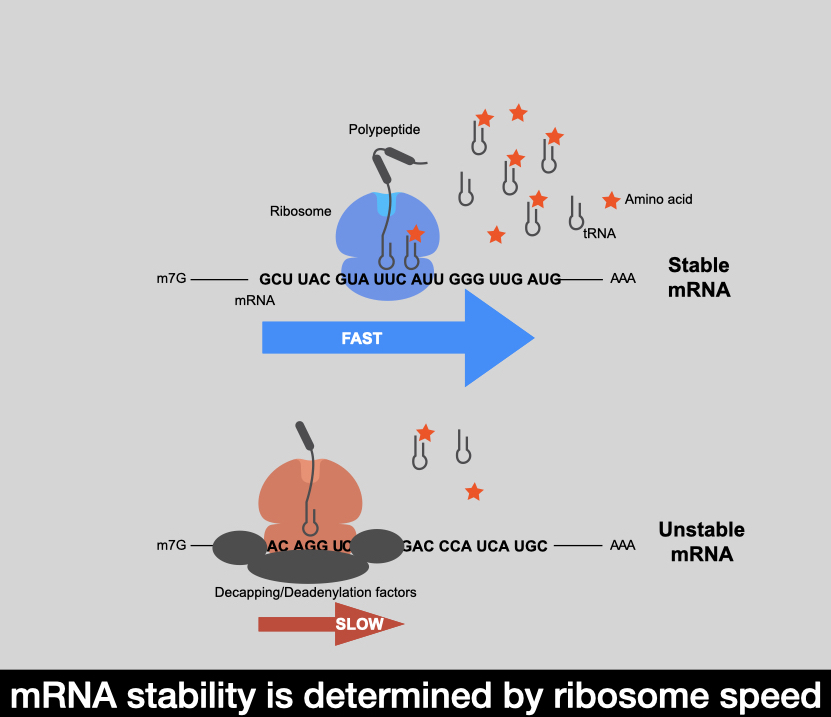
Messenger RNA (mRNA) degradation plays a critical role in regulating transcript levels in the cell and is a major control point for modulating gene expression. mRNA stability is as vital as transcription in determining steady state RNA levels; but it is comparatively less understood. For over 40 years, it has been appreciated that in dividing cells, mRNA turnover is tightly coupled to the translation status of the message. Specifically, mRNA that is efficiently translated is more stable than mRNA that is poorly translated. The nature of this relationship has been a mystery. My lab has studied the interrelationship between mRNA translation and mRNA stability as a platform to investigate how mRNA half-lives are determined. A series of discoveries from my group have uncovered that a significant determinant for RNA degradation that substantially contributes to differential half-lives is the genetic code itself and a concept termed “Codon Optimality”. Specifically, we have shown that mRNA decay rates are dictated by the percentage of codons deemed ‘optimal’ (based on the abundance of their cognate tRNAs relative to demand). We hypothesized that optimal codons are decoded by ribosomes more rapidly, while non-optimal codons are read more slowly (due to limiting tRNA concentration and base-pairing thermostability. Thus the rate at which the ribosome decodes a mRNA ultimately determines transcript fate. Now we study how regulation is built into this system by looking at 1] how the mRNA degradation machinery interacts with the ribosome 2] how features such as RNA modifications can impact codon optimality and 3] how tRNA biology can ultimately impact mRNA levels. Lastly, we are harnessing these discoveries, pushing open this exciting field, and leveraging our findings towards the development of novel therapeutics.
MORE DETAILS
Messenger RNA (mRNA) translation by the ribosome represents the final step on a complicated molecular dance from DNA to protein. Though classically considered a decipherer that translates our 64-word genetic code into a proteome of astonishing complexity, recent work has unexpectedly shown that the ribosome can also shape the expressed mRNA transcriptome. General mRNA degradation occurs stepwise with removal of the 3’ poly(A) tail (deadenylation), removal of the 5’ m7GpppN cap (decapping), and degradation of the transcript body 5’ to 3’ by exonuclease. We recently discovered that the ribosome is a master arbiter of this general mRNA degradation pathway, wherein ribosome transit rate serves as a major determinant of transcript half-lives. Specifically, we found that members of the degradation complex sense ribosome translocation rate as a function of codon composition. Central to this notion is the concept of Codon Optimality: though all codons impact translation rate, some are deciphered quickly while others cause ribosome hesitation (a consequence of relative cognate tRNA concentration). It is our hypothesis that these transient pauses induce a unique ribosome conformational state that is probed by the degradation complex, thereby inducing an orchestrated set of events that enhance both deadenylation and decapping rates (see figure below). Together, these data implicate that the coding region of an mRNA not only encodes for protein content, but also impacts protein levels through determining the transcript’s fate.
My lab has a history of producing paradigm shifting work. We leverage both yeast and mammalian systems to study the processes of mRNA translation and mRNA stability. We have uncovered a major mechanism that broadly determines mRNA half-lives, revealing a new and exciting area of exploration. Next, we will define the molecular interplay between the ribosome and the degradation complex in response to codon optimality. Most excitingly is our observation that the exact same mRNA sequence can differentially impact mRNA stability in distinct cell types. Thus, codon optimality is plastic in nature and is regulated from cell-to-cell. Preliminary data points toward two features governing cell-specific codon optimality: differential tRNA expression and differential mRNA modification. Lastly, our work highlights a powerful and manipulable way to artificially control mRNA expression. We have modulated codon optimality, and associated mRNA levels, through the expression of specific tRNA isodecoders. This represents an exciting opportunity through which fundamental research can inform on therapeutics that would be useful, for example, in the treatment of diseases stemming from insufficient protein, such as haploinsufficiency disorders. A unifying principle from our work is that codon optimality is a fundamental property governing mRNA stability that can be regulated and manipulated to achieve gene regulation.
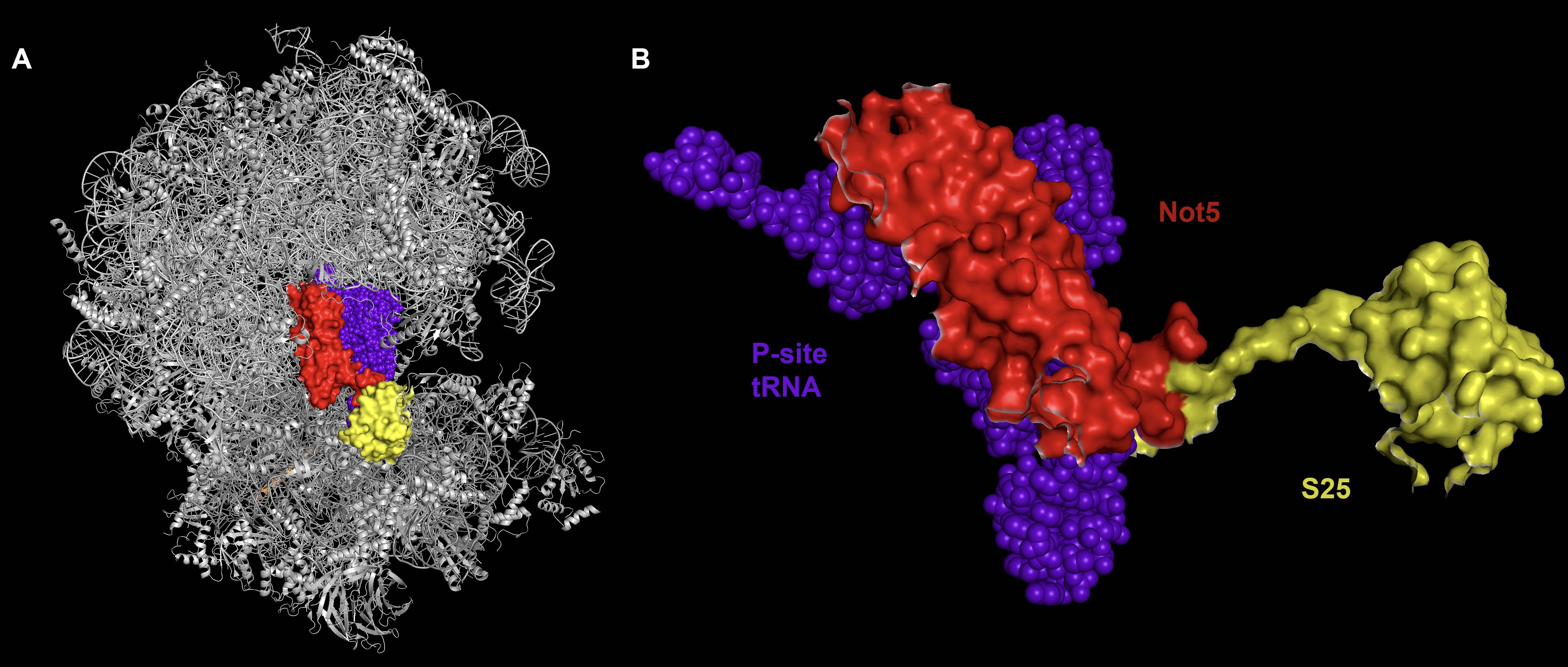
Show is a cryo-EM structure of the ribosome. In response to poor codon optimality, the decapping protein, NOT5 (CNOT3 in humans) probes the ribosome E-site and touchs the P-site tRNA. The mechanistic consquence of E-site probing by the decapping complex is under investigation.